R Markdown
This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see http://rmarkdown.rstudio.com.
When you click the Knit button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this:
####Eggs PER PLANT ####
Sub.Eggs <- subset(mydata,
select=c(Nger,
Isolate,
EggsP) )
##Create a subset of the desire variableEggs per plant: MIAd
####Isolate: MIAd####
Sub.Eggs.MIAd <- subset(Sub.Eggs, Isolate == "MIAd", select=c(Nger, EggsP))
Sub.Eggs.MIAd$Nger = factor (Sub.Eggs.MIAd$Nger, labels=c("EC", "TB", "TE", "TS", "TT"))
###Descriptive statistics calculation###
Sub.Eggs.MIAd.EC <- subset(Sub.Eggs.MIAd, Nger == "EC", select=c(Nger,EggsP))
Me.EP.MIAd.EC <- mean(Sub.Eggs.MIAd.EC$EggsP, na.mr=FALSE)
Me.EP.MIAd.EC <- round(Me.EP.MIAd.EC, digits=2)
Se.EP.MIAd.EC <- se(Sub.Eggs.MIAd.EC$EggsP)
Se.EP.MIAd.EC <- round(Se.EP.MIAd.EC, digits=2)
Sub.Eggs.MIAd.SB <- subset(Sub.Eggs.MIAd, Nger == "TB", select=c(Nger,EggsP))
Me.EP.MIAd.TB <- mean(Sub.Eggs.MIAd.SB$EggsP, na.mr=FALSE)
Me.EP.MIAd.TB <- round(Me.EP.MIAd.TB, digits=2)
Se.EP.MIAd.TB <- se(Sub.Eggs.MIAd.SB$EggsP)
Se.EP.MIAd.TB <- round(Se.EP.MIAd.TB, digits=2)
Sub.Eggs.MIAd.SB <- subset(Sub.Eggs.MIAd, Nger == "TE", select=c(Nger,EggsP))
Me.EP.MIAd.TE <- mean(Sub.Eggs.MIAd.SB$EggsP, na.mr=FALSE)
Me.EP.MIAd.TE <- round(Me.EP.MIAd.TE, digits=2)
Se.EP.MIAd.TE <- se(Sub.Eggs.MIAd.SB$EggsP)
Se.EP.MIAd.TE <- round(Se.EP.MIAd.TE, digits=2)
Sub.Eggs.MIAd.SB <- subset(Sub.Eggs.MIAd, Nger == "TS", select=c(Nger,EggsP))
Me.EP.MIAd.TS <- mean(Sub.Eggs.MIAd.SB$EggsP, na.mr=FALSE)
Me.EP.MIAd.TS <- round(Me.EP.MIAd.TS, digits=2)
Se.EP.MIAd.TS <- se(Sub.Eggs.MIAd.SB$EggsP)
Se.EP.MIAd.TS <- round(Se.EP.MIAd.TS, digits=2)
Sub.Eggs.MIAd.SB <- subset(Sub.Eggs.MIAd, Nger == "TT", select=c(Nger,EggsP))
Me.EP.MIAd.TT <- mean(Sub.Eggs.MIAd.SB$EggsP, na.mr=FALSE)
Me.EP.MIAd.TT <- round(Me.EP.MIAd.TT, digits=2)
Se.EP.MIAd.TT <- se(Sub.Eggs.MIAd.SB$EggsP)
Se.EP.MIAd.TT <- round(Se.EP.MIAd.TT, digits=2)
####Kruskall-Wallis test calculation####
KTMIAd = kruskal.test(Sub.Eggs.MIAd$EggsP ~ Sub.Eggs.MIAd$Nger)
####Dunn test calculation####
DTMIAd = dunnTest(Sub.Eggs.MIAd$EggsP ~ Sub.Eggs.MIAd$Nger, method="bonferroni")
DTMIAd.Comparison <- DTMIAd$res$Comparison
DTMIAd.Padj <- round (DTMIAd$res$P.adj, digits=5)
KrusData = data.frame(DTMIAd.Comparison, DTMIAd.Padj)
RND = nrow(DTMIAd$res)
krustable=kable(KrusData[1:RND,])Descriptive statistics
| Germplasm | Mean ± SE | Dif |
|---|---|---|
| EC: | 31517 ± 7491 | |
| TB: | 65 ± 25 | |
| TE: | 360 ± 106 | |
| TS: | 1328 ± 538 | |
| TT: | 220 ± 45 |
Non parametic test
Dunn (1964) Kruskal-Wallis multiple comparison p-values adjusted with the Bonferroni method:
if (KTMIAd$p.value>0.05) {
dif="There is no significative differences (P<0.05) between treatments (P<0.05)"
krustable="0"
} else {
dif="There is significative differences between treatments (P<0.05)."
krustable=kable(KrusData[1:RND,])
}There is significative differences between treatments (P<0.05).
Including Plots
You can also embed plots, for example:
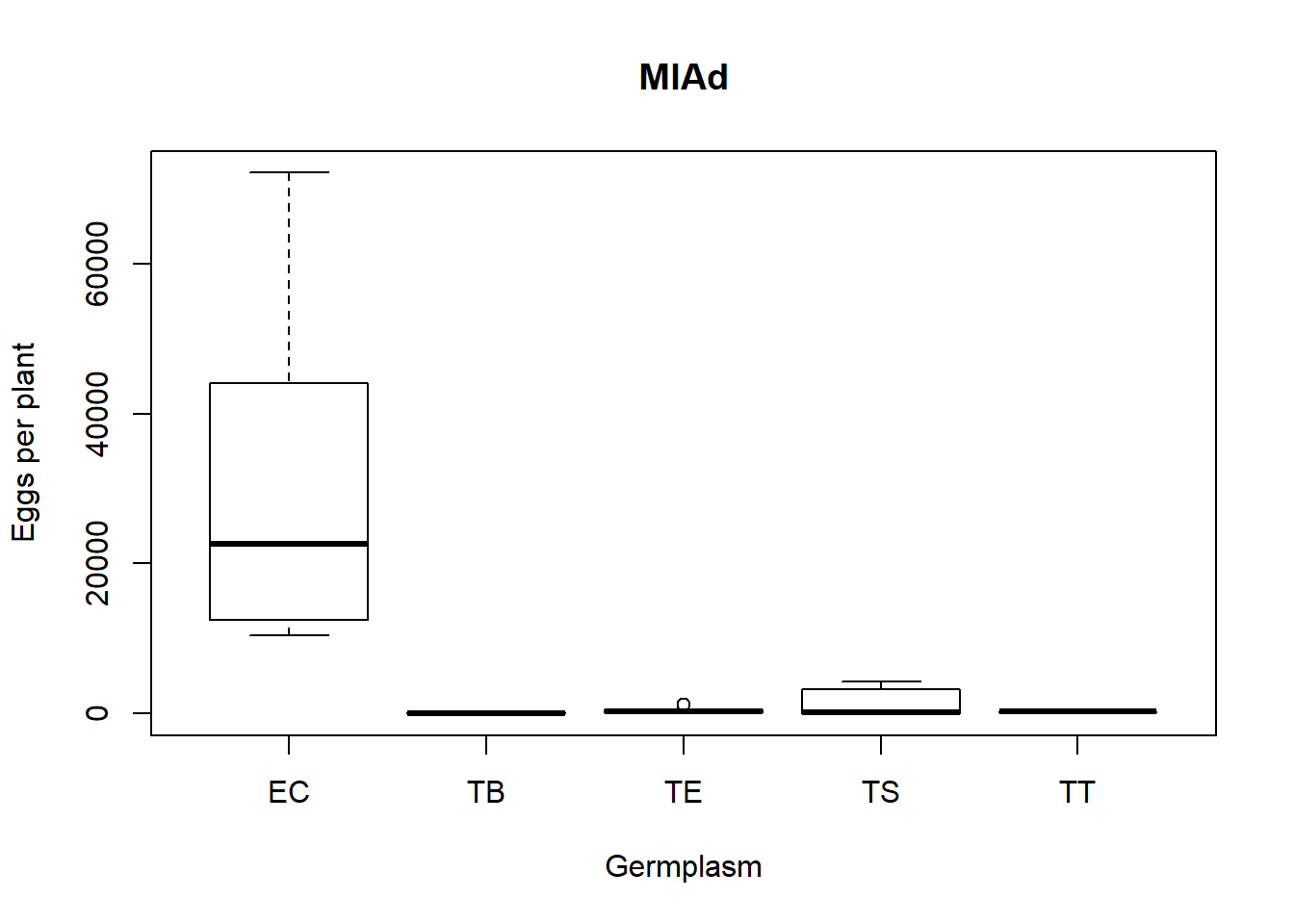
Note that the echo = FALSE parameter was added to the code chunk to prevent printing of the R code that generated the plot.
Dunn (1964) Kruskal-Wallis multiple comparison p-values adjusted with the Bonferroni method.
DTMIAd$dtres## [1] " Kruskal-Wallis rank sum test"
## [2] ""
## [3] "data: x and g"
## [4] "Kruskal-Wallis chi-squared = 29.3877, df = 4, p-value = 0"
## [5] ""
## [6] ""
## [7] " Comparison of x by g "
## [8] " (Bonferroni) "
## [9] "Col Mean-|"
## [10] "Row Mean | EC TB TE TS"
## [11] "---------+--------------------------------------------"
## [12] " TB | 5.264338"
## [13] " | 0.0000*"
## [14] " |"
## [15] " TE | 3.020269 -2.244068"
## [16] " | 0.0253* 0.2483"
## [17] " |"
## [18] " TS | 3.496750 -1.767587 0.476480"
## [19] " | 0.0047* 0.7713 1.0000"
## [20] " |"
## [21] " TT | 3.588972 -1.675365 0.568702 0.092221"
## [22] " | 0.0033* 0.9386 1.0000 1.0000"
## [23] ""
## [24] "alpha = 0"
## [25] "Reject Ho if p <= alpha"